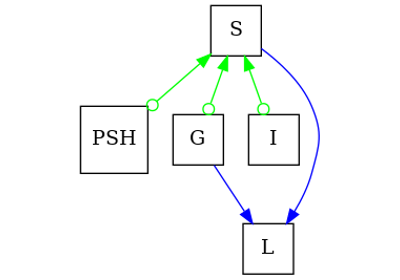
pywhy_graphs.networkx.MixedEdgeGraph#
- class pywhy_graphs.networkx.MixedEdgeGraph(graphs=None, edge_types=None, **attr)[source]#
Base class for mixed-edge graphs.
A mixed-edge graph stores nodes and different kinds of edges. The edges can represent non-directed (i.e.
networkx.Graph), or directed (i.e.networkx.DiGraph) edge connections among nodes. Nodes can be any nodes that can be represented innetworkx.Graph, andnetworkx.DiGraph.Edges are represented as links between nodes with optional key/value attributes.
Warning
MixedEdgeGraph is an experimental feature that is meant to be included into networkx eventually. Please use with caution.
- Parameters:
- graphsList of Graph | DiGraph
A list of networkx single-edge graphs.
- edge_typesList of str
A list of names for each edge type.
- attrkeyword arguments, optional (default= no attributes)
Attributes to add to graph as key=value pairs.
Notes
Besides the changes mentioned below, a
MixedEdgeGraphmatches the entire API provided by the Graph/DiGraph classes.Changes compared to existing networkx graphs:
Compared to
networkx.Graphandnetworkx.DiGraph, aMixedEdgeGraphhas a different method for initializing the graph and adding edges. When adding/removing/update edges to the graph, if that edge type does not exist, then an error will be raised. Users should explicitly add an edge type graph via theadd_edge_typefunction.Moreover, computing an
edge_subgraphis not supported forMixedEdgeGraph.Edges, Adjacencies and Degree:
Compared to single-edge networkx graphs,
MixedEdgeGraphimplements these as functions rather than cached properties.Neighbors: Compared to single-edge graphs, neighbors in a
MixedEdgeGraphof a node is defined as any other node with an edge connected to the node that is being checked.Keywords: Since
MixedEdgeGraphcomprises of possibly many different edge types. In the API, there are a few keywords: ‘any’ or ‘all’. Within the API, ‘any’ means any edge type, whereas ‘all’ refers to all edge types.- __getitem__(n)[source]#
Returns a dict of neighbors of node n. Use: ‘G[n]’.
The adjacency dictionary for nodes connected to n.
Notes
G[n] is the same as G.adj[n] and similar to G.neighbors(n) (which is an iterator over G.adj[n])
- __iter__()[source]#
Iterate over the nodes. Use: ‘for n in G’.
An iterator over all nodes in the graph.
- __len__()[source]#
Returns the number of nodes in the graph. Use: ‘len(G)’.
See also
number_of_nodesidentical method
orderidentical method
- add_edge(u_of_edge, v_of_edge, edge_type='all', **attr)[source]#
Add an edge between u and v.
The nodes u and v will be automatically added if they are not already in the graph.
Edge attributes can be specified with keywords or by directly accessing the edge’s attribute dictionary.
- Parameters:
- u_for_edge, v_for_edgenodes
Nodes can be, for example, strings or numbers. Nodes must be hashable (and not None) Python objects.
- edge_typestr
The edge type. By default ‘all’, which will then add an edge in all edge type subgraphs.
- attrkeyword arguments, optional
Edge data (or labels or objects) can be assigned using keyword arguments.
See also
add_edges_fromadd a collection of edges
- add_edges_from(ebunch_to_add, edge_type, **attr)[source]#
Add all the edges in ebunch_to_add.
- Parameters:
- ebunch_to_addcontainer of edges
Each edge given in the container will be added to the graph. The edges must be given as 2-tuples (u, v) or 3-tuples (u, v, d) where d is a dictionary containing edge data.
- edge_typestr
The edge type to add edges to. By default ‘all’, which will then add an edge in all edge type subgraphs.
- attrkeyword arguments, optional
Edge data (or labels or objects) can be assigned using keyword arguments.
See also
add_edgeadd a single edge
Notes
Adding the same edge twice has no effect but any edge data will be updated when each duplicate edge is added.
Edge attributes specified in an ebunch take precedence over attributes specified via keyword arguments.
- property adj#
Dictionary of graph adjacency objects holding the neighbors of each node.
Each edge type has an adjacency object associated with it. For more information on the adjacency object itself, see the documentation in
networkx.Graph.adj.Iterating over G.adj behaves like a dict. Useful idioms include the following for loop.
>>> for edge_type, adj in G.adj.items(): >>> for nbr, datadict in adj[n].items(): >>> ...
The main difference from non-mixed edge graph types is that
adjhere returns a dictionary of adjacency views, so neighbors can be queried within each edge type.- Returns:
- adjdictionary of AdjacencyView
A dictionary of edge types and their corresponding adjacency view objects.
- copy()[source]#
Returns a copy of the graph.
The copy method by default returns an independent shallow copy of the graph and attributes. That is, if an attribute is a container, that container is shared by the original an the copy. Use Python’s
copy.deepcopyfor new containers.- Parameters:
- as_viewbool, optional (default=False)
If True, the returned graph-view provides a read-only view of the original graph without actually copying any data.
- Returns:
- GGraph
A copy of the graph.
See also
to_directedreturn a directed copy of the graph.
networkx.Graph.copyfor full copy docstring.
Notes
All copies reproduce the graph structure, but data attributes may be handled in different ways. There are four types of copies of a graph that people might want. See
networkx.Graph.copy()for doc string.
- degree(nbunch=None, weight=None)[source]#
A DegreeView for the Graph as G.degree or G.degree().
- Parameters:
- nbunchsingle node, container, or all nodes (default= all nodes)
The view will only report edges incident to these nodes.
- weightstring or None, optional (default=None)
The name of an edge attribute that holds the numerical value used as a weight. If None, then each edge has weight 1. The degree is the sum of the edge weights adjacent to the node.
- Returns:
- deg_dictsdictionary of DegreeView | int
If multiple nodes are requested (the default), returns a
DegreeViewmapping nodes to their degree. If a single node is requested, returns the degree of the node as an integer.
- edges(nbunch=None, data=False, default=None)[source]#
A dictionary of EdgeViews of the Graph as G.edges or G.edges().
Each edge type has an EdgeView object associated with it. For more information on the EdgeView object itself, see the documentation in
networkx.Graph.edges.- Parameters:
- nbunchsingle node, container, or all nodes (default= all nodes)
The view will only report edges from these nodes.
- datastring or bool, optional (default=False)
The edge attribute returned in 3-tuple (u, v, ddict[data]). If True, return edge attribute dict in 3-tuple (u, v, ddict). If False, return 2-tuple (u, v).
- defaultvalue, optional (default=None)
Value used for edges that don’t have the requested attribute. Only relevant if data is not True or False.
- Returns:
- edgesdictionary of EdgeView
A dictionary of EdgeViews.
Notes
Nodes in nbunch that are not in the graph will be (quietly) ignored. For directed graphs this returns the out-edges.
- get_edge_data(u, v, default=None)[source]#
Returns the attribute dictionary associated with edge (u, v).
This is identical to
G[u][v]except the default is returned instead of an exception if the edge doesn’t exist.- Parameters:
- u, vnodes
- default: any Python object (default=None)
Value to return if the edge (u, v) is not found.
- Returns:
- edge_dictdictionary
The edge attribute dictionary.
- get_graphs(edge_type='all') Graph | Dict[str, Graph][source]#
Get graphs representing the mixed-edges.
- Parameters:
- edge_typestr, optional
The graph of the edge type to return, by default ‘all’, which will then return a dictionary of all edge graphs keyed by their edge type string.
- Returns:
- graphGraph | dictionary of Graph
The graph representing a specific type of edge, or all edges.
- has_edge(u, v, edge_type='any')[source]#
Returns True if the edge (u, v) is in the graph.
This is the same as
v in G[u]without KeyError exceptions.- Parameters:
- u, vnodes
Nodes can be, for example, strings or numbers. Nodes must be hashable (and not None) Python objects.
- edge_typestr
Specifies a specific edge type. If ‘any’ (default), then will check if any edge exists between
uandvacross all edge types.
- Returns:
- edge_indbool
True if edge is in the graph, False otherwise.
- has_node(n)[source]#
Returns True if the graph contains the node n.
Identical to
n in G- Parameters:
- nnode
The node to query. Identical to
n in G.
- Returns:
- hasbool
Whether or not the graph has node ‘n’.
- property name#
String identifier of the graph.
This graph attribute appears in the attribute dict G.graph keyed by the string
"name". as well as an attribute (technically a property)G.name. This is entirely user controlled.
- nbunch_iter(nbunch=None)[source]#
Returns an iterator over nodes contained in nbunch that are also in the graph.
The nodes in nbunch are checked for membership in the graph and if not are silently ignored.
- Parameters:
- nbunchsingle node, container, or all nodes (default= all nodes)
The view will only report edges incident to these nodes.
- Returns:
- niteriterator
An iterator over nodes in nbunch that are also in the graph. If nbunch is None, iterate over all nodes in the graph.
- Raises:
- NetworkXError
If nbunch is not a node or sequence of nodes. If a node in nbunch is not hashable.
- number_of_edges(u=None, v=None, edge_type='all')[source]#
Returns the number of edges between two nodes.
- Parameters:
- u, vnodes, optional (default=all edges)
If u and v are specified, return the number of edges between u and v. Otherwise return the total number of all edges.
- edge_typestr, optional
The edge type to query for number of edges. If ‘all’ (default) will count the number of edges regardless of edge type.
- Returns:
- nedgesint
The number of edges in the graph. If nodes
uandvare specified return the number of edges between those nodes. If the graph is directed, this only returns the number of edges fromutov.
See also
- order()[source]#
Returns the number of nodes in the graph.
See also
number_of_nodesidentical method
__len__identical method
- remove_edge(u, v, edge_type='all')[source]#
Remove an edge between u and v.
- Parameters:
- u, vnodes
Remove an edge between nodes u and v.
- edge_typestr
The edge type. By default ‘all’, which will then remove the edge in all edge type subgraphs (if it exists).
- Raises:
- NetworkXError
If there is not an edge between u and v, or if there is no edge with the specified key.
See also
remove_edges_fromremove a collection of edges
- remove_edges_from(ebunch, edge_type='all')[source]#
Remove all edges specified in ebunch.
- Parameters:
- ebunch: list or container of edge tuples
Each edge given in the list or container will be removed from the graph. The edges can be:
2-tuples (u, v) edge between u and v.
3-tuples (u, v, k) where k is ignored.
See also
remove_edgeremove a single edge
Notes
Will fail silently if an edge in ebunch is not in the graph.
- size(weight=None, edge_type='all')[source]#
Returns the number of edges or total of all edge weights.
- Parameters:
- weightstring or None, optional (default=None)
The edge attribute that holds the numerical value used as a weight. If None, then each edge has weight 1.
- Returns:
- sizefloat
The number of edges or (if weight keyword is provided) the total weight sum. If weight is None, returns an int. Otherwise a float (or more general numeric if the weights are more general).
See also
- subgraph(nodes)[source]#
Returns a SubGraph view of the subgraph induced on
nodes.TODO: this does not work as expected with subclasses The induced subgraph of the graph contains the nodes in
nodesand the edges between those nodes.- Parameters:
- nodeslist, iterable
A container of nodes which will be iterated through once.
- Returns:
- GMixedEdgeGraph
A copy of the graph with only the nodes.
- to_directed()[source]#
Returns a directed representation of the graph.
- Returns:
- GDiGraph
A directed graph with the same name, same nodes, and with each edge (u, v, data) replaced by two directed edges (u, v, data) and (v, u, data).
Notes
This returns a “deepcopy” of the edge, node, and graph attributes which attempts to completely copy all of the data and references.
This is in contrast to the similar D=DiGraph(G) which returns a shallow copy of the data.
See the Python copy module for more information on shallow and deep copies, https://docs.python.org/3/library/copy.html.
Warning: If you have subclassed Graph to use dict-like objects in the data structure, those changes do not transfer to the DiGraph created by this method.
- to_undirected()[source]#
Returns an undirected representation of the digraph.
- Returns:
- GGraph
An undirected graph with the same name and nodes and with edge (u, v, data) if either (u, v, data) or (v, u, data) is in the digraph. If both edges exist in a sub digraph and their edge data is different, only one edge is created with an arbitrary choice of which edge data to use. You must check and correct for this manually if desired.
Notes
This returns a “deepcopy” of the edge, node, and graph attributes which attempts to completely copy all of the data and references.
This is in contrast to the similar D=MultiDiGraph(G) which returns a shallow copy of the data.
See the Python copy module for more information on shallow and deep copies, https://docs.python.org/3/library/copy.html.
Warning: If you have subclassed MultiDiGraph to use dict-like objects in the data structure, those changes do not transfer to the MultiGraph created by this method.
- update(edges=None, nodes=None, edge_type=None)[source]#
Update the graph using nodes/edges/graphs as input.
Like dict.update, this method takes a graph as input, adding the graph’s nodes and edges to this graph. It can also take two inputs: edges and nodes. Finally it can take either edges or nodes. To specify only nodes the keyword
nodesmust be used.The collections of edges and nodes are treated similarly to the add_edges_from/add_nodes_from methods. When iterated, they should yield 2-tuples (u, v) or 3-tuples (u, v, datadict).
- Parameters:
- edgesnx.Graph object, collection of edges, or None
The first parameter can be a graph or some edges. If it has attributes
nodesandedges, then it is taken to be a Graph-like object and those attributes are used as collections of nodes and edges to be added to the graph. If the first parameter does not have those attributes, it is treated as a collection of edges and added to the graph. If the first argument is None, no edges are added.- nodescollection of nodes, or None
The second parameter is treated as a collection of nodes to be added to the graph unless it is None. If
edges is Noneandnodes is Nonean exception is raised. If the first parameter is a Graph, thennodesis ignored.
Examples using pywhy_graphs.networkx.MixedEdgeGraph#
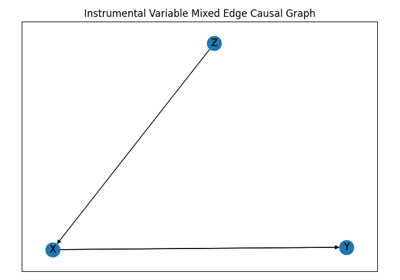
MixedEdgeGraph - Graph with different types of edges
An introduction to Inducing Paths and how to find them
An introduction to causal graphs and how to use them
Drawing graphs and setting their layout for visual comparison