pywhy_graphs.PAG#
- class pywhy_graphs.PAG(incoming_directed_edges=None, incoming_undirected_edges=None, incoming_bidirected_edges=None, incoming_circle_edges=None, directed_edge_name: str = 'directed', undirected_edge_name: str = 'undirected', bidirected_edge_name: str = 'bidirected', circle_edge_name: str = 'circle', **attr)[source]#
Partial ancestral graph (PAG).
PAGs are a Markov equivalence class with mixed edges of directed, bidirected, undirected and edges with circle endpoints. In terms of graph functionality, they essentially extend the definition of an ADMG with edges with circular endpoints.
- Parameters:
- incoming_directed_edgesinput directed edges (optional, default: None)
Data to initialize directed edges. All arguments that are accepted by
networkx.DiGraphare accepted.- incoming_undirected_edgesinput undirected edges (optional, default: None)
Data to initialize undirected edges. All arguments that are accepted by
networkx.Graphare accepted.- incoming_bidirected_edgesinput bidirected edges (optional, default: None)
Data to initialize bidirected edges. All arguments that are accepted by
networkx.Graphare accepted.- incoming_circle_edgesinput circular endpoint edges (optional, default: None)
Data to initialize edges with circle endpoints. All arguments that are accepted by
networkx.DiGraphare accepted.- directed_edge_namestr
The name for the directed edges. By default ‘directed’.
- undirected_edge_namestr
The name for the undirected edges. By default ‘undirected’.
- bidirected_edge_namestr
The name for the bidirected edges. By default ‘bidirected’.
- circle_edge_namestr
The name for the circle edges. By default ‘circle’.
- attrkeyword arguments, optional (default= no attributes)
Attributes to add to graph as key=value pairs.
Notes
PAGs are Markov equivalence class of causal ADMGs. The implicit assumption in these causal graphs are the Structural Causal Model (or SCM) is Semi-Markovian, such that latent confounders may be present. This allows PAGs to be learned from constraint-based (such as the FCI algorithm) approaches for causal structure learning.
Edge Type Subgraphs
The data structure underneath the hood is stored in two types of networkx graphs:
networkx.Graphandnetworkx.DiGraph.- Directed edges (<-, ->, indicating causal relationship) =
networkx.DiGraph The subgraph of directed edges may be accessed by the
~.PAG.sub_directed_graph. Their edges in networkx format can be accessed bydirected_edgesand the corresponding name of the edge type bydirected_edge_name.
- Directed edges (<-, ->, indicating causal relationship) =
- Bidirected edges (<->, indicating latent confounder) =
networkx.Graph The subgraph of bidirected edges may be accessed by the
~.PAG.sub_bidirected_graph. Their edges in networkx format can be accessed bybidirected_edgesand the corresponding name of the edge type bybidirected_edge_name.
- Bidirected edges (<->, indicating latent confounder) =
- Undirected edges (–, indicating selection bias) =
networkx.Graph The subgraph of undirected edges may be accessed by the
~.PAG.sub_undirected_graph. Their edges in networkx format can be accessed by~.PAG.undirected_edgesand the corresponding name of the edge type by~.PAG.undirected_edge_name.
- Undirected edges (–, indicating selection bias) =
- Circle edges (-o, o-, indicating uncertainty) =
networkx.DiGraph The subgraph of undirected edges may be accessed by the
~.PAG.sub_circle_graph. Their edges in networkx format can be accessed by~.PAG.circle_edgesand the corresponding name of the edge type by~.PAG.circle_edge_name.
- Circle edges (-o, o-, indicating uncertainty) =
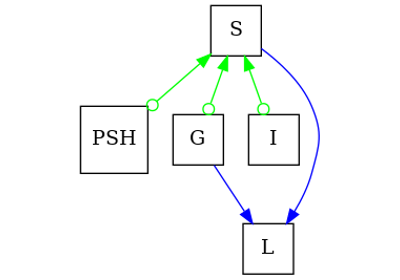
How different edges are represented in the PAG
Compared to an
~pywhy_graphs.classes.ADMGand~pywhy_graphs.classes.CPDAGand anetworkx.DiGraph, a PAG is more complex in that it generalizes endpoints an edge can take, exponentially increasing the number of possible edges that can occur between two nodes. The main complication arises in edges with circle endpoints. Rather than store all possible edges as separate networkx graphs, we have a set of rules that map a combination of the above edge-type subgraphs to a certain edge.Bidirected and undirected edges are represented by one networkx graph (
networkx.Graph). They are simple in that they do not require pairing with another edge-type subgraph.x <-> y: is a bidirected edge present? (Note by definition of a PAG no other edgecan be present between x and y)
x - y: is an undirected present? (Note no other edge should be present in anyother direction, so an undirected edge is similar to a bidirected edge in that it represents only one kind of edge)
Edges with arrowheads, tails and circular endpoints are represented by another networkx graph (
networkx.DiGraph). They complicate matters because the~.PAG.sub_directed_graphand~.PAG.sub_circle_graphcan be combined in different ways to result in different edges between x and y.Without loss of generality, we will be dealing with the ordered tuple (x, y). If you want the other direction of the edge, you can just flip the order of x and y. For example,
x <- ywould just bey -> x, so we will only discuss the->edge. The following rules dictate what sort of edge we are dealing with:x o-o y: is circle edge present in both directions? There are only edges presentin the
~.PAG.sub_circle_graphbetween x and y.
x o-> y: is circle edge one way and directed edge another way? There is an edge fromthe
~.PAG.sub_circle_graphand the~.PAG.sub_directed_graphbetween x and y in opposite directions.
x o- y: is there only one circle edge? In this special case, we do not use the~.PAG.sub_undirected_graphto represent the tail endpoint at y. There is only one edge in the~.PAG.sub_circle_graphbetween x and y.
- add_edge(u_of_edge, v_of_edge, edge_type='all', **attr)[source]#
Add an edge between u and v.
The nodes u and v will be automatically added if they are not already in the graph.
Edge attributes can be specified with keywords or by directly accessing the edge’s attribute dictionary.
- Parameters:
- u_for_edge, v_for_edgenodes
Nodes can be, for example, strings or numbers. Nodes must be hashable (and not None) Python objects.
- edge_typestr
The edge type. By default ‘all’, which will then add an edge in all edge type subgraphs.
- attrkeyword arguments, optional
Edge data (or labels or objects) can be assigned using keyword arguments.
See also
add_edges_fromadd a collection of edges
- add_edges_from(ebunch_to_add, edge_type, **attr)[source]#
Add all the edges in ebunch_to_add.
- Parameters:
- ebunch_to_addcontainer of edges
Each edge given in the container will be added to the graph. The edges must be given as 2-tuples (u, v) or 3-tuples (u, v, d) where d is a dictionary containing edge data.
- edge_typestr
The edge type to add edges to. By default ‘all’, which will then add an edge in all edge type subgraphs.
- attrkeyword arguments, optional
Edge data (or labels or objects) can be assigned using keyword arguments.
See also
add_edgeadd a single edge
Notes
Adding the same edge twice has no effect but any edge data will be updated when each duplicate edge is added.
Edge attributes specified in an ebunch take precedence over attributes specified via keyword arguments.
- children(n: int | float | str | Any) Iterator[source]#
Return the definite children of node ‘n’ in a PAG.
Definite children are children of node ‘n’ with only a directed edge between them from ‘n’ -> ‘x’. For example, ‘n’ o-> ‘x’ does not qualify ‘x’ as a children of ‘n’.
- Parameters:
- nnode
A node in the causal DAG.
- Yields:
- childrenIterator
An iterator of the children of node ‘n’.
See also
- orient_uncertain_edge(u: int | float | str | Any, v: int | float | str | Any) None[source]#
Orient undirected edge into an arrowhead.
If there is an undirected edge u - v, then the arrowhead will orient u -> v. If the correct order is v <- u, then simply pass the arguments in different order.
- Parameters:
- unode
The parent node
- vnode
The node that ‘u’ points to in the graph.
- parents(n: int | float | str | Any) Iterator[source]#
Return the definite parents of node ‘n’ in a PAG.
Definite parents are parents of node ‘n’ with only a directed edge between them from ‘n’ <- ‘x’. For example, ‘n’ <-o ‘x’ does not qualify ‘x’ as a parent of ‘n’.
- Parameters:
- nnode
A node in the causal DAG.
- Yields:
- parentsIterator
An iterator of the definite parents of node ‘n’.
See also
- possible_children(n: int | float | str | Any) Iterator[source]#
Return an iterator over children of node n.
Possible children of ‘n’ are nodes with an edge like
'n' o-> 'x'. Nodes with'n' <-* 'x'are not considered possible children.- Parameters:
- nnode
A node in the causal DAG.
- Returns:
- possible_childrenIterator
An iterator of the children of node ‘n’.
- possible_parents(n: int | float | str | Any) Iterator[source]#
Return an iterator over possible parents of node n.
Possible parents of ‘n’ are nodes with an edge like
'n' <-* 'x'. Nodes with'n' *-> 'x'are not considered possible parents.- Parameters:
- nnode
A node in the causal DAG.
- Returns:
- possible_parentsIterator
An iterator of the parents of node ‘n’.
Examples using pywhy_graphs.PAG#
An introduction to causal graphs and how to use them
Drawing graphs and setting their layout for visual comparison