Note
Go to the end to download the full example code.
On PAGs and their validity#
A PAG or a Partial Ancestral Graph is a type of mixed edge graph that can represent, in a single graph, the causal relationship between several nodes as defined by an equivalence class of MAGs.
PAGs account for possible unobserved confounding and selection bias in the underlying equivalence class of SCMs.
Another way to understand this is that PAGs encode conditional independence constraints stemming from Causal Graphs. Since these constraints do not lead to a unique graph, a PAG, in essence, represents a class of graphs that encode the same conditional independence constraints.
PAGs model this relationship by displaying all common edge marks (tail and arrowhead) shared
by all members in the equivalence class and displaying circle endpoints for those marks
that are not common. That is, a circular endpoint (*-o) can represent both a directed
(*->) and tail (*—) endpoint in causal graphs within the equivalence class.
More details on PAGs can be found at [1].
import pywhy_graphs
from pywhy_graphs.viz import draw
from pywhy_graphs import PAG
try:
from dodiscover import FCI, make_context
from dodiscover.ci import Oracle
from dodiscover.constraint.utils import dummy_sample
except ImportError as e:
raise ImportError("The 'dodiscover' package is required to convert a MAG to a PAG.")
PAGs in pywhy-graphs#
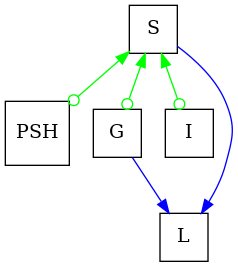
Constructing a PAG in pywhy-graphs is an easy task since the library provides a separate class for this purpose. True to the definition of PAGs, the class can contain directed edges, bidirected edges, undirected edges and cicle edges. To illustrate this, we construct an example PAG as described in [1], figure 4:
pag = PAG()
pag.add_edge("I", "S", pag.directed_edge_name)
pag.add_edge("G", "S", pag.directed_edge_name)
pag.add_edge("G", "L", pag.directed_edge_name)
pag.add_edge("S", "L", pag.directed_edge_name)
pag.add_edge("PSH", "S", pag.directed_edge_name)
pag.add_edge("S", "PSH", pag.circle_edge_name)
pag.add_edge("S", "G", pag.circle_edge_name)
pag.add_edge("S", "I", pag.circle_edge_name)
# Finally, the graph looks like this:
dot_graph = draw(pag)
dot_graph.render(outfile="valid_pag.png", view=True)
'valid_pag.png'
Validity of a PAG#
For a PAG to be valid, it must represent a valid equivalent class of MAGs. This can be verified by turning the PAG into an MAG and then checking the validity of the MAG. Theorem 2 in [1] provides a method for checking the validity of a PAG. To check if the constructed PAG is a valid one in pywhy-graphs, we can simply do:
# returns True
print(pywhy_graphs.valid_pag(pag))
ConditioningSetSelection.PDS
Context(observed_variables={'PSH', 'S', 'L', 'I', 'G'}, latent_variables=set(), state_variables={}, init_graph=<networkx.classes.graph.Graph object at 0x7f7a50e27110>, included_edges=<networkx.classes.graph.Graph object at 0x7f7a50e27810>, excluded_edges=<networkx.classes.graph.Graph object at 0x7f7a50e27c50>, num_distributions=1, obs_distribution=True, intervention_targets=[], symmetric_diff_map={}, sigma_map={}, f_nodes=[], num_domains=1, domain_map={}, s_nodes=[])
True
If we want to test whether this algorithm is working correctly or not, we can change
a single mark in the graph such that the PAG. By removing a circle edge, we are removing
the representation of multiple marks as encoded by the different MAGs this PAG represents.
In this specific case, by removing the circle endpoint S *-o I, we are saying that S
directly causes I. However, there is no way of determining this using the FCI logical rules.
One would not be able to determine that the adjacency is due to a direct
causal relationship (directed edge), confounded relationship (bidirected edge), or an inducing path
relationship. As such, the resulting graph is no longer a valid PAG.
pag.remove_edge("S", "I", pag.circle_edge_name)
# returns False
print(pywhy_graphs.valid_pag(pag))
ConditioningSetSelection.PDS
Context(observed_variables={'PSH', 'S', 'L', 'I', 'G'}, latent_variables=set(), state_variables={}, init_graph=<networkx.classes.graph.Graph object at 0x7f7a50e375d0>, included_edges=<networkx.classes.graph.Graph object at 0x7f7a50e37cd0>, excluded_edges=<networkx.classes.graph.Graph object at 0x7f7a50e44150>, num_distributions=1, obs_distribution=True, intervention_targets=[], symmetric_diff_map={}, sigma_map={}, f_nodes=[], num_domains=1, domain_map={}, s_nodes=[])
False
References#
Total running time of the script: (0 minutes 1.092 seconds)
Estimated memory usage: 167 MB