Demo for the DoWhy causal API#
We show a simple example of adding a causal extension to any dataframe.
[1]:
import dowhy.datasets
import dowhy.api
from dowhy.graph import build_graph_from_str
import numpy as np
import pandas as pd
from statsmodels.api import OLS
[2]:
data = dowhy.datasets.linear_dataset(beta=5,
num_common_causes=1,
num_instruments = 0,
num_samples=1000,
treatment_is_binary=True)
df = data['df']
df['y'] = df['y'] + np.random.normal(size=len(df)) # Adding noise to data. Without noise, the variance in Y|X, Z is zero, and mcmc fails.
nx_graph = build_graph_from_str(data["dot_graph"])
treatment= data["treatment_name"][0]
outcome = data["outcome_name"][0]
common_cause = data["common_causes_names"][0]
df
[2]:
| W0 | v0 | y | |
|---|---|---|---|
| 0 | 0.037488 | False | -0.174667 |
| 1 | -0.523050 | False | -1.292853 |
| 2 | 2.070420 | True | 9.443895 |
| 3 | 0.068183 | False | 1.113386 |
| 4 | 0.104074 | True | 5.843495 |
| ... | ... | ... | ... |
| 995 | -0.265148 | True | 4.197523 |
| 996 | -0.973204 | False | -0.142424 |
| 997 | -1.241690 | False | -2.795862 |
| 998 | -1.825721 | False | -3.752621 |
| 999 | -0.607617 | True | 2.775188 |
1000 rows × 3 columns
[3]:
# data['df'] is just a regular pandas.DataFrame
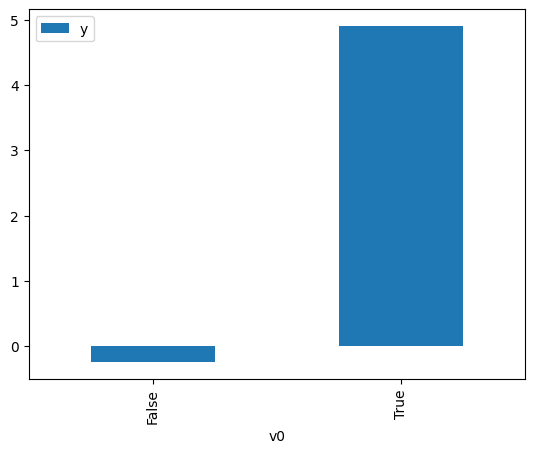
df.causal.do(x=treatment,
variable_types={treatment: 'b', outcome: 'c', common_cause: 'c'},
outcome=outcome,
common_causes=[common_cause],
).groupby(treatment).mean().plot(y=outcome, kind='bar')
[3]:
<Axes: xlabel='v0'>
[4]:
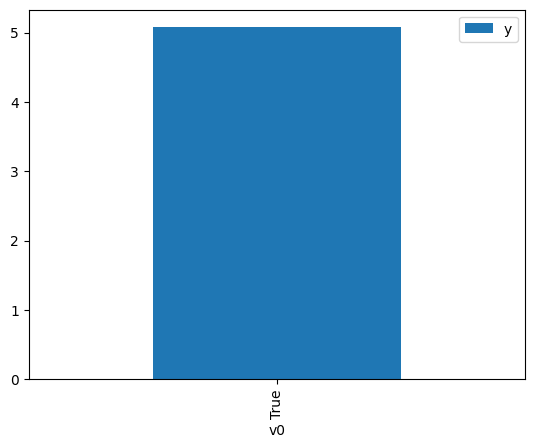
df.causal.do(x={treatment: 1},
variable_types={treatment:'b', outcome: 'c', common_cause: 'c'},
outcome=outcome,
method='weighting',
common_causes=[common_cause]
).groupby(treatment).mean().plot(y=outcome, kind='bar')
[4]:
<Axes: xlabel='v0'>
[5]:
cdf_1 = df.causal.do(x={treatment: 1},
variable_types={treatment: 'b', outcome: 'c', common_cause: 'c'},
outcome=outcome,
graph=nx_graph
)
cdf_0 = df.causal.do(x={treatment: 0},
variable_types={treatment: 'b', outcome: 'c', common_cause: 'c'},
outcome=outcome,
graph=nx_graph
)
[6]:
cdf_0
[6]:
| W0 | v0 | y | propensity_score | weight | |
|---|---|---|---|---|---|
| 0 | -0.713942 | False | -0.832839 | 0.774908 | 1.290476 |
| 1 | 0.378506 | False | 1.757012 | 0.329282 | 3.036907 |
| 2 | -0.416285 | False | -1.897409 | 0.669419 | 1.493834 |
| 3 | -1.022786 | False | -3.304443 | 0.856541 | 1.167486 |
| 4 | 0.541459 | False | 1.429673 | 0.268558 | 3.723594 |
| ... | ... | ... | ... | ... | ... |
| 995 | -0.873678 | False | 0.280653 | 0.820688 | 1.218489 |
| 996 | 0.933586 | False | 2.296218 | 0.154328 | 6.479718 |
| 997 | -0.961088 | False | -1.653206 | 0.842488 | 1.186961 |
| 998 | -0.057126 | False | 0.769879 | 0.516302 | 1.936850 |
| 999 | -0.328091 | False | 0.223023 | 0.633746 | 1.577919 |
1000 rows × 5 columns
[7]:
cdf_1
[7]:
| W0 | v0 | y | propensity_score | weight | |
|---|---|---|---|---|---|
| 0 | -0.752117 | True | 3.312724 | 0.213443 | 4.685087 |
| 1 | -0.568538 | True | 4.378100 | 0.273487 | 3.656476 |
| 2 | 1.378671 | True | 7.617855 | 0.923760 | 1.082532 |
| 3 | 0.917073 | True | 6.610054 | 0.841791 | 1.187943 |
| 4 | -0.871327 | True | 2.153620 | 0.179929 | 5.557742 |
| ... | ... | ... | ... | ... | ... |
| 995 | 0.260021 | True | 5.172035 | 0.622504 | 1.606415 |
| 996 | 3.495361 | True | 14.133175 | 0.998108 | 1.001895 |
| 997 | -1.526145 | True | 1.050194 | 0.063908 | 15.647445 |
| 998 | -0.324753 | True | 4.160558 | 0.367636 | 2.720078 |
| 999 | 1.709055 | True | 6.392574 | 0.956211 | 1.045794 |
1000 rows × 5 columns
Comparing the estimate to Linear Regression#
First, estimating the effect using the causal data frame, and the 95% confidence interval.
[8]:
(cdf_1['y'] - cdf_0['y']).mean()
[8]:
$\displaystyle 5.40925954703069$
[9]:
1.96*(cdf_1['y'] - cdf_0['y']).std() / np.sqrt(len(df))
[9]:
$\displaystyle 0.191657653401046$
Comparing to the estimate from OLS.
[10]:
model = OLS(np.asarray(df[outcome]), np.asarray(df[[common_cause, treatment]], dtype=np.float64))
result = model.fit()
result.summary()
[10]:
| Dep. Variable: | y | R-squared (uncentered): | 0.957 |
|---|---|---|---|
| Model: | OLS | Adj. R-squared (uncentered): | 0.957 |
| Method: | Least Squares | F-statistic: | 1.102e+04 |
| Date: | Sun, 24 Nov 2024 | Prob (F-statistic): | 0.00 |
| Time: | 18:03:52 | Log-Likelihood: | -1410.0 |
| No. Observations: | 1000 | AIC: | 2824. |
| Df Residuals: | 998 | BIC: | 2834. |
| Df Model: | 2 | ||
| Covariance Type: | nonrobust |
| coef | std err | t | P>|t| | [0.025 | 0.975] | |
|---|---|---|---|---|---|---|
| x1 | 2.1425 | 0.034 | 63.535 | 0.000 | 2.076 | 2.209 |
| x2 | 4.9804 | 0.048 | 103.470 | 0.000 | 4.886 | 5.075 |
| Omnibus: | 0.734 | Durbin-Watson: | 1.928 |
|---|---|---|---|
| Prob(Omnibus): | 0.693 | Jarque-Bera (JB): | 0.658 |
| Skew: | 0.060 | Prob(JB): | 0.720 |
| Kurtosis: | 3.038 | Cond. No. | 1.67 |
Notes:
[1] R² is computed without centering (uncentered) since the model does not contain a constant.
[2] Standard Errors assume that the covariance matrix of the errors is correctly specified.