Demo for the DoWhy causal API
We show a simple example of adding a causal extension to any dataframe.
[1]:
import dowhy.datasets
import dowhy.api
from dowhy.graph import build_graph_from_str
import numpy as np
import pandas as pd
from statsmodels.api import OLS
[2]:
data = dowhy.datasets.linear_dataset(beta=5,
num_common_causes=1,
num_instruments = 0,
num_samples=1000,
treatment_is_binary=True)
df = data['df']
df['y'] = df['y'] + np.random.normal(size=len(df)) # Adding noise to data. Without noise, the variance in Y|X, Z is zero, and mcmc fails.
nx_graph = build_graph_from_str(data["dot_graph"])
treatment= data["treatment_name"][0]
outcome = data["outcome_name"][0]
common_cause = data["common_causes_names"][0]
df
[2]:
| W0 | v0 | y | |
|---|---|---|---|
| 0 | -0.330622 | False | 0.887729 |
| 1 | -0.691849 | False | -1.118670 |
| 2 | 0.193915 | False | -0.966624 |
| 3 | -0.535931 | True | 2.642023 |
| 4 | 2.334936 | True | 7.962068 |
| ... | ... | ... | ... |
| 995 | 0.339981 | False | -0.948675 |
| 996 | -0.919134 | False | 0.069790 |
| 997 | -1.140944 | False | -0.716580 |
| 998 | 0.352853 | True | 4.586724 |
| 999 | -0.983083 | True | 6.705248 |
1000 rows × 3 columns
[3]:
# data['df'] is just a regular pandas.DataFrame
df.causal.do(x=treatment,
variable_types={treatment: 'b', outcome: 'c', common_cause: 'c'},
outcome=outcome,
common_causes=[common_cause],
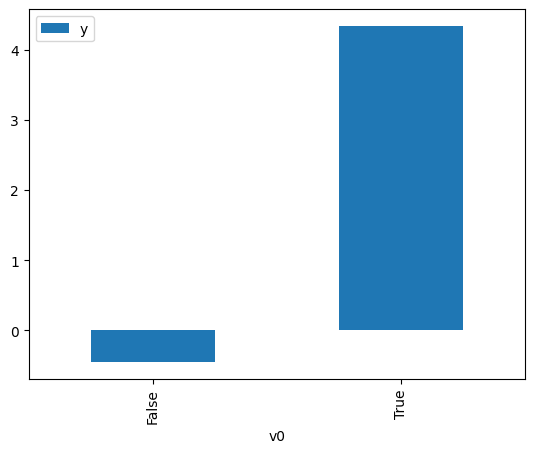
).groupby(treatment).mean().plot(y=outcome, kind='bar')
[3]:
<Axes: xlabel='v0'>
[4]:
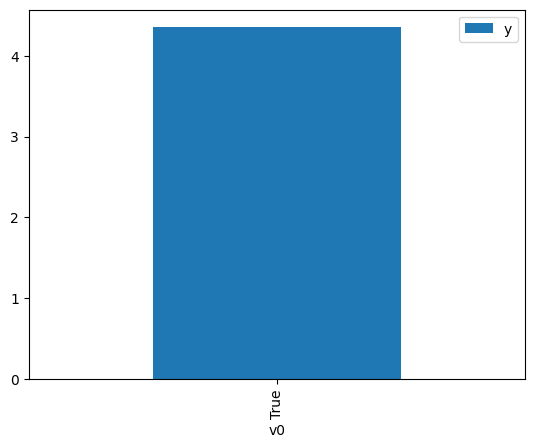
df.causal.do(x={treatment: 1},
variable_types={treatment:'b', outcome: 'c', common_cause: 'c'},
outcome=outcome,
method='weighting',
common_causes=[common_cause]
).groupby(treatment).mean().plot(y=outcome, kind='bar')
[4]:
<Axes: xlabel='v0'>
[5]:
cdf_1 = df.causal.do(x={treatment: 1},
variable_types={treatment: 'b', outcome: 'c', common_cause: 'c'},
outcome=outcome,
graph=nx_graph
)
cdf_0 = df.causal.do(x={treatment: 0},
variable_types={treatment: 'b', outcome: 'c', common_cause: 'c'},
outcome=outcome,
graph=nx_graph
)
[6]:
cdf_0
[6]:
| W0 | v0 | y | propensity_score | weight | |
|---|---|---|---|---|---|
| 0 | -0.416171 | False | -1.103170 | 0.587029 | 1.703494 |
| 1 | 1.020597 | False | 1.610446 | 0.256853 | 3.893284 |
| 2 | 0.289512 | False | 1.261112 | 0.415118 | 2.408955 |
| 3 | -0.807504 | False | -1.508775 | 0.676307 | 1.478619 |
| 4 | -0.127578 | False | 0.523608 | 0.516908 | 1.934579 |
| ... | ... | ... | ... | ... | ... |
| 995 | 0.193915 | False | -0.966624 | 0.438129 | 2.282434 |
| 996 | -0.663361 | False | -1.209901 | 0.644508 | 1.551571 |
| 997 | 0.507649 | False | -0.926918 | 0.364117 | 2.746373 |
| 998 | 1.261393 | False | 0.781840 | 0.214268 | 4.667049 |
| 999 | -1.187408 | False | -1.447086 | 0.752270 | 1.329309 |
1000 rows × 5 columns
[7]:
cdf_1
[7]:
| W0 | v0 | y | propensity_score | weight | |
|---|---|---|---|---|---|
| 0 | -1.096630 | True | 1.579037 | 0.264752 | 3.777119 |
| 1 | 0.883869 | True | 6.774463 | 0.716633 | 1.395414 |
| 2 | -1.335124 | True | 2.590650 | 0.221639 | 4.511850 |
| 3 | -1.064736 | True | 4.180466 | 0.270908 | 3.691296 |
| 4 | -0.712749 | True | 2.574438 | 0.344435 | 2.903306 |
| ... | ... | ... | ... | ... | ... |
| 995 | -0.394069 | True | 4.852584 | 0.418255 | 2.390889 |
| 996 | -2.109866 | True | -0.132723 | 0.117258 | 8.528192 |
| 997 | 1.342959 | True | 7.022089 | 0.798937 | 1.251663 |
| 998 | 0.263952 | True | 3.750980 | 0.578761 | 1.727828 |
| 999 | -1.284152 | True | 2.070227 | 0.230414 | 4.340014 |
1000 rows × 5 columns
Comparing the estimate to Linear Regression
First, estimating the effect using the causal data frame, and the 95% confidence interval.
[8]:
(cdf_1['y'] - cdf_0['y']).mean()
[8]:
$\displaystyle 4.86323421622536$
[9]:
1.96*(cdf_1['y'] - cdf_0['y']).std() / np.sqrt(len(df))
[9]:
$\displaystyle 0.141690908498881$
Comparing to the estimate from OLS.
[10]:
model = OLS(np.asarray(df[outcome]), np.asarray(df[[common_cause, treatment]], dtype=np.float64))
result = model.fit()
result.summary()
[10]:
| Dep. Variable: | y | R-squared (uncentered): | 0.921 |
|---|---|---|---|
| Model: | OLS | Adj. R-squared (uncentered): | 0.921 |
| Method: | Least Squares | F-statistic: | 5840. |
| Date: | Wed, 06 Dec 2023 | Prob (F-statistic): | 0.00 |
| Time: | 08:43:32 | Log-Likelihood: | -1436.2 |
| No. Observations: | 1000 | AIC: | 2876. |
| Df Residuals: | 998 | BIC: | 2886. |
| Df Model: | 2 | ||
| Covariance Type: | nonrobust |
| coef | std err | t | P>|t| | [0.025 | 0.975] | |
|---|---|---|---|---|---|---|
| x1 | 1.1002 | 0.028 | 39.374 | 0.000 | 1.045 | 1.155 |
| x2 | 4.9739 | 0.050 | 99.704 | 0.000 | 4.876 | 5.072 |
| Omnibus: | 0.374 | Durbin-Watson: | 1.968 |
|---|---|---|---|
| Prob(Omnibus): | 0.829 | Jarque-Bera (JB): | 0.380 |
| Skew: | 0.047 | Prob(JB): | 0.827 |
| Kurtosis: | 2.984 | Cond. No. | 1.79 |
Notes:
[1] R² is computed without centering (uncentered) since the model does not contain a constant.
[2] Standard Errors assume that the covariance matrix of the errors is correctly specified.