Demo for the DoWhy causal API
We show a simple example of adding a causal extension to any dataframe.
[1]:
import dowhy.datasets
import dowhy.api
import numpy as np
import pandas as pd
from statsmodels.api import OLS
[2]:
data = dowhy.datasets.linear_dataset(beta=5,
num_common_causes=1,
num_instruments = 0,
num_samples=1000,
treatment_is_binary=True)
df = data['df']
df['y'] = df['y'] + np.random.normal(size=len(df)) # Adding noise to data. Without noise, the variance in Y|X, Z is zero, and mcmc fails.
#data['dot_graph'] = 'digraph { v ->y;X0-> v;X0-> y;}'
treatment= data["treatment_name"][0]
outcome = data["outcome_name"][0]
common_cause = data["common_causes_names"][0]
df
[2]:
| W0 | v0 | y | |
|---|---|---|---|
| 0 | -1.085634 | False | -4.084557 |
| 1 | -1.418167 | False | -4.159834 |
| 2 | -1.060908 | False | -1.876372 |
| 3 | -0.040109 | True | 4.739495 |
| 4 | -1.135199 | False | -2.126015 |
| ... | ... | ... | ... |
| 995 | 0.167694 | True | 6.002419 |
| 996 | -1.342202 | False | -5.061231 |
| 997 | -1.115815 | False | -4.549248 |
| 998 | 0.224101 | False | 2.319660 |
| 999 | -0.559218 | False | -1.882604 |
1000 rows × 3 columns
[3]:
# data['df'] is just a regular pandas.DataFrame
df.causal.do(x=treatment,
variable_types={treatment: 'b', outcome: 'c', common_cause: 'c'},
outcome=outcome,
common_causes=[common_cause],
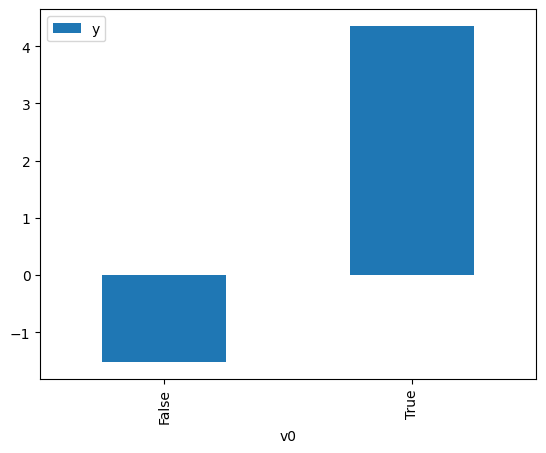
proceed_when_unidentifiable=True).groupby(treatment).mean().plot(y=outcome, kind='bar')
[3]:
<Axes: xlabel='v0'>
[4]:
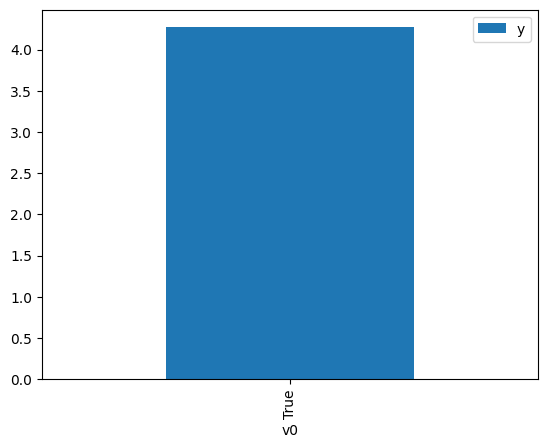
df.causal.do(x={treatment: 1},
variable_types={treatment:'b', outcome: 'c', common_cause: 'c'},
outcome=outcome,
method='weighting',
common_causes=[common_cause],
proceed_when_unidentifiable=True).groupby(treatment).mean().plot(y=outcome, kind='bar')
[4]:
<Axes: xlabel='v0'>
[5]:
cdf_1 = df.causal.do(x={treatment: 1},
variable_types={treatment: 'b', outcome: 'c', common_cause: 'c'},
outcome=outcome,
dot_graph=data['dot_graph'],
proceed_when_unidentifiable=True)
cdf_0 = df.causal.do(x={treatment: 0},
variable_types={treatment: 'b', outcome: 'c', common_cause: 'c'},
outcome=outcome,
dot_graph=data['dot_graph'],
proceed_when_unidentifiable=True)
[6]:
cdf_0
[6]:
| W0 | v0 | y | propensity_score | weight | |
|---|---|---|---|---|---|
| 0 | -0.550093 | False | -1.532310 | 0.756892 | 1.321192 |
| 1 | -0.722684 | False | -1.846021 | 0.810431 | 1.233912 |
| 2 | -1.996264 | False | -6.116133 | 0.977962 | 1.022534 |
| 3 | -0.525880 | False | -0.314164 | 0.748614 | 1.335802 |
| 4 | -0.668037 | False | -3.078179 | 0.794523 | 1.258616 |
| ... | ... | ... | ... | ... | ... |
| 995 | -2.188685 | False | -5.719837 | 0.984423 | 1.015824 |
| 996 | 0.607096 | False | 0.420697 | 0.270844 | 3.692155 |
| 997 | -0.310138 | False | -0.762520 | 0.667047 | 1.499146 |
| 998 | -1.141617 | False | -4.197842 | 0.902252 | 1.108338 |
| 999 | 0.757894 | False | 1.761406 | 0.219703 | 4.551601 |
1000 rows × 5 columns
[7]:
cdf_1
[7]:
| W0 | v0 | y | propensity_score | weight | |
|---|---|---|---|---|---|
| 0 | -0.568209 | True | 3.579572 | 0.237036 | 4.218773 |
| 1 | -0.506941 | True | 3.209480 | 0.257991 | 3.876099 |
| 2 | 0.701984 | True | 7.612798 | 0.762181 | 1.312024 |
| 3 | -0.486111 | True | 4.661744 | 0.265385 | 3.768110 |
| 4 | 0.228094 | True | 5.046634 | 0.572979 | 1.745264 |
| ... | ... | ... | ... | ... | ... |
| 995 | -0.472540 | True | 2.449277 | 0.270274 | 3.699946 |
| 996 | -1.012103 | True | 1.651103 | 0.120835 | 8.275773 |
| 997 | -1.472749 | True | 0.877186 | 0.055678 | 17.960356 |
| 998 | -0.103875 | True | 3.710211 | 0.421676 | 2.371490 |
| 999 | -0.375534 | True | 5.820682 | 0.306825 | 3.259191 |
1000 rows × 5 columns
Comparing the estimate to Linear Regression
First, estimating the effect using the causal data frame, and the 95% confidence interval.
[8]:
(cdf_1['y'] - cdf_0['y']).mean()
[8]:
$\displaystyle 5.45773159896272$
[9]:
1.96*(cdf_1['y'] - cdf_0['y']).std() / np.sqrt(len(df))
[9]:
$\displaystyle 0.247766786137291$
Comparing to the estimate from OLS.
[10]:
model = OLS(np.asarray(df[outcome]), np.asarray(df[[common_cause, treatment]], dtype=np.float64))
result = model.fit()
result.summary()
[10]:
| Dep. Variable: | y | R-squared (uncentered): | 0.955 |
|---|---|---|---|
| Model: | OLS | Adj. R-squared (uncentered): | 0.955 |
| Method: | Least Squares | F-statistic: | 1.062e+04 |
| Date: | Thu, 03 Aug 2023 | Prob (F-statistic): | 0.00 |
| Time: | 18:29:21 | Log-Likelihood: | -1426.4 |
| No. Observations: | 1000 | AIC: | 2857. |
| Df Residuals: | 998 | BIC: | 2867. |
| Df Model: | 2 | ||
| Covariance Type: | nonrobust |
| coef | std err | t | P>|t| | [0.025 | 0.975] | |
|---|---|---|---|---|---|---|
| x1 | 2.7773 | 0.030 | 93.411 | 0.000 | 2.719 | 2.836 |
| x2 | 5.0002 | 0.054 | 91.951 | 0.000 | 4.894 | 5.107 |
| Omnibus: | 2.397 | Durbin-Watson: | 1.975 |
|---|---|---|---|
| Prob(Omnibus): | 0.302 | Jarque-Bera (JB): | 2.464 |
| Skew: | -0.105 | Prob(JB): | 0.292 |
| Kurtosis: | 2.878 | Cond. No. | 1.89 |
Notes:
[1] R² is computed without centering (uncentered) since the model does not contain a constant.
[2] Standard Errors assume that the covariance matrix of the errors is correctly specified.