Demo for the DoWhy causal API
We show a simple example of adding a causal extension to any dataframe.
[1]:
import dowhy.datasets
import dowhy.api
import numpy as np
import pandas as pd
from statsmodels.api import OLS
[2]:
data = dowhy.datasets.linear_dataset(beta=5,
num_common_causes=1,
num_instruments = 0,
num_samples=1000,
treatment_is_binary=True)
df = data['df']
df['y'] = df['y'] + np.random.normal(size=len(df)) # Adding noise to data. Without noise, the variance in Y|X, Z is zero, and mcmc fails.
#data['dot_graph'] = 'digraph { v ->y;X0-> v;X0-> y;}'
treatment= data["treatment_name"][0]
outcome = data["outcome_name"][0]
common_cause = data["common_causes_names"][0]
df
[2]:
| W0 | v0 | y | |
|---|---|---|---|
| 0 | 0.615341 | False | -0.216582 |
| 1 | 1.547448 | True | 3.791882 |
| 2 | -1.220144 | True | 4.319986 |
| 3 | -0.167506 | True | 6.049082 |
| 4 | 2.950866 | True | 4.455041 |
| ... | ... | ... | ... |
| 995 | 0.978907 | True | 6.964400 |
| 996 | 2.768105 | True | 4.851054 |
| 997 | 0.657292 | True | 7.053682 |
| 998 | 0.462598 | False | 0.075282 |
| 999 | 0.544589 | True | 3.206106 |
1000 rows × 3 columns
[3]:
# data['df'] is just a regular pandas.DataFrame
df.causal.do(x=treatment,
variable_types={treatment: 'b', outcome: 'c', common_cause: 'c'},
outcome=outcome,
common_causes=[common_cause],
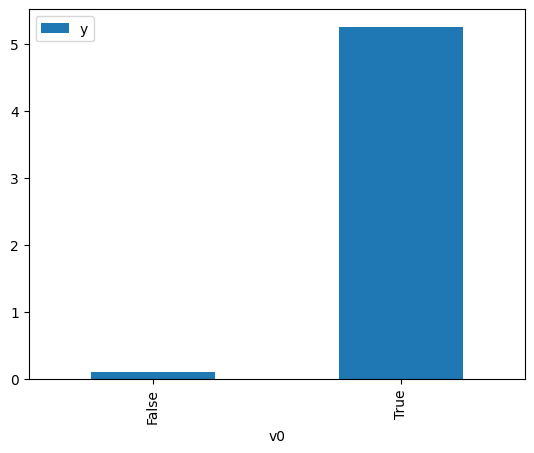
proceed_when_unidentifiable=True).groupby(treatment).mean().plot(y=outcome, kind='bar')
[3]:
<AxesSubplot: xlabel='v0'>
[4]:
df.causal.do(x={treatment: 1},
variable_types={treatment:'b', outcome: 'c', common_cause: 'c'},
outcome=outcome,
method='weighting',
common_causes=[common_cause],
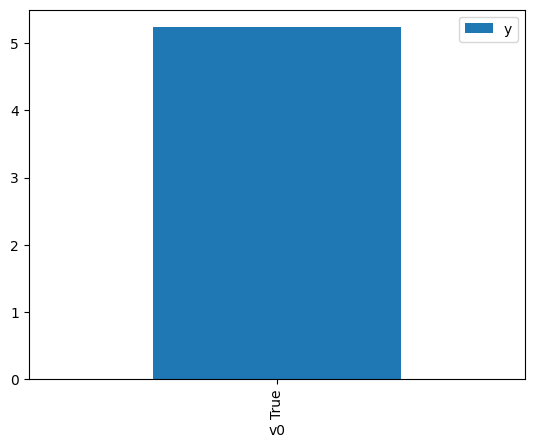
proceed_when_unidentifiable=True).groupby(treatment).mean().plot(y=outcome, kind='bar')
[4]:
<AxesSubplot: xlabel='v0'>
[5]:
cdf_1 = df.causal.do(x={treatment: 1},
variable_types={treatment: 'b', outcome: 'c', common_cause: 'c'},
outcome=outcome,
dot_graph=data['dot_graph'],
proceed_when_unidentifiable=True)
cdf_0 = df.causal.do(x={treatment: 0},
variable_types={treatment: 'b', outcome: 'c', common_cause: 'c'},
outcome=outcome,
dot_graph=data['dot_graph'],
proceed_when_unidentifiable=True)
[6]:
cdf_0
[6]:
| W0 | v0 | y | propensity_score | weight | |
|---|---|---|---|---|---|
| 0 | 0.067560 | False | 1.911379 | 0.466482 | 2.143707 |
| 1 | 0.823216 | False | -1.139482 | 0.298868 | 3.345962 |
| 2 | 1.662468 | False | 0.487500 | 0.161029 | 6.210075 |
| 3 | 1.219407 | False | -0.734138 | 0.226293 | 4.419058 |
| 4 | 3.170608 | False | 0.352740 | 0.043754 | 22.854844 |
| ... | ... | ... | ... | ... | ... |
| 995 | 2.290390 | False | 0.137876 | 0.095559 | 10.464770 |
| 996 | 1.037646 | False | 0.709049 | 0.257967 | 3.876460 |
| 997 | -0.134015 | False | 1.177118 | 0.514338 | 1.944246 |
| 998 | 0.438583 | False | -2.028024 | 0.380597 | 2.627451 |
| 999 | -0.018765 | False | -1.847223 | 0.486952 | 2.053589 |
1000 rows × 5 columns
[7]:
cdf_1
[7]:
| W0 | v0 | y | propensity_score | weight | |
|---|---|---|---|---|---|
| 0 | 0.151752 | True | 5.144520 | 0.553375 | 1.807092 |
| 1 | -0.361275 | True | 5.457729 | 0.432065 | 2.314466 |
| 2 | -0.516995 | True | 4.685548 | 0.396163 | 2.524211 |
| 3 | -0.151791 | True | 7.376667 | 0.481441 | 2.077096 |
| 4 | 0.533555 | True | 4.762663 | 0.640448 | 1.561408 |
| ... | ... | ... | ... | ... | ... |
| 995 | 2.328699 | True | 5.015506 | 0.907543 | 1.101876 |
| 996 | 0.922553 | True | 6.789926 | 0.720539 | 1.387849 |
| 997 | 0.890339 | True | 5.813846 | 0.714331 | 1.399912 |
| 998 | -0.380071 | True | 4.574668 | 0.427686 | 2.338166 |
| 999 | 1.219794 | True | 3.804645 | 0.773772 | 1.292371 |
1000 rows × 5 columns
Comparing the estimate to Linear Regression
First, estimating the effect using the causal data frame, and the 95% confidence interval.
[8]:
(cdf_1['y'] - cdf_0['y']).mean()
[8]:
$\displaystyle 5.18094679145088$
[9]:
1.96*(cdf_1['y'] - cdf_0['y']).std() / np.sqrt(len(df))
[9]:
$\displaystyle 0.0932881847772126$
Comparing to the estimate from OLS.
[10]:
model = OLS(np.asarray(df[outcome]), np.asarray(df[[common_cause, treatment]], dtype=np.float64))
result = model.fit()
result.summary()
[10]:
| Dep. Variable: | y | R-squared (uncentered): | 0.945 |
|---|---|---|---|
| Model: | OLS | Adj. R-squared (uncentered): | 0.945 |
| Method: | Least Squares | F-statistic: | 8643. |
| Date: | Tue, 06 Dec 2022 | Prob (F-statistic): | 0.00 |
| Time: | 09:38:55 | Log-Likelihood: | -1450.1 |
| No. Observations: | 1000 | AIC: | 2904. |
| Df Residuals: | 998 | BIC: | 2914. |
| Df Model: | 2 | ||
| Covariance Type: | nonrobust |
| coef | std err | t | P>|t| | [0.025 | 0.975] | |
|---|---|---|---|---|---|---|
| x1 | 0.2364 | 0.035 | 6.773 | 0.000 | 0.168 | 0.305 |
| x2 | 5.0849 | 0.053 | 95.597 | 0.000 | 4.981 | 5.189 |
| Omnibus: | 0.226 | Durbin-Watson: | 1.922 |
|---|---|---|---|
| Prob(Omnibus): | 0.893 | Jarque-Bera (JB): | 0.264 |
| Skew: | 0.035 | Prob(JB): | 0.876 |
| Kurtosis: | 2.962 | Cond. No. | 2.46 |
Notes:
[1] R² is computed without centering (uncentered) since the model does not contain a constant.
[2] Standard Errors assume that the covariance matrix of the errors is correctly specified.