Demo for the DoWhy causal API
We show a simple example of adding a causal extension to any dataframe.
[1]:
import dowhy.datasets
import dowhy.api
from dowhy.graph import build_graph_from_str
import numpy as np
import pandas as pd
from statsmodels.api import OLS
[2]:
data = dowhy.datasets.linear_dataset(beta=5,
num_common_causes=1,
num_instruments = 0,
num_samples=1000,
treatment_is_binary=True)
df = data['df']
df['y'] = df['y'] + np.random.normal(size=len(df)) # Adding noise to data. Without noise, the variance in Y|X, Z is zero, and mcmc fails.
nx_graph = build_graph_from_str(data["dot_graph"])
treatment= data["treatment_name"][0]
outcome = data["outcome_name"][0]
common_cause = data["common_causes_names"][0]
df
[2]:
| W0 | v0 | y | |
|---|---|---|---|
| 0 | -1.352371 | False | -2.800845 |
| 1 | 1.840101 | True | 5.312718 |
| 2 | 1.462809 | True | 5.135988 |
| 3 | -1.268049 | False | -0.090203 |
| 4 | -1.062236 | False | 1.062694 |
| ... | ... | ... | ... |
| 995 | 0.278762 | False | 0.058603 |
| 996 | 1.429788 | True | 9.476343 |
| 997 | -2.136987 | False | -2.413670 |
| 998 | -2.070323 | False | -1.066857 |
| 999 | -0.415496 | True | 3.013789 |
1000 rows × 3 columns
[3]:
# data['df'] is just a regular pandas.DataFrame
df.causal.do(x=treatment,
variable_types={treatment: 'b', outcome: 'c', common_cause: 'c'},
outcome=outcome,
common_causes=[common_cause],
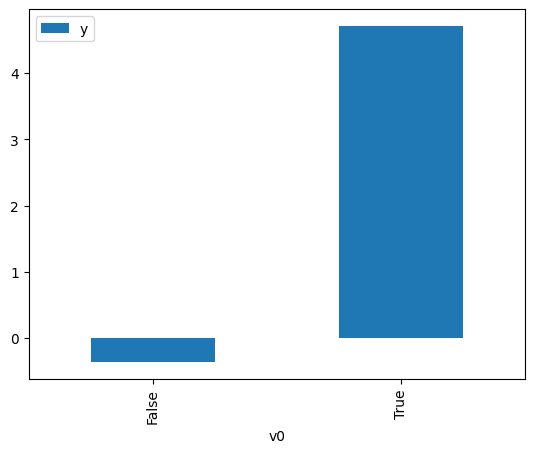
).groupby(treatment).mean().plot(y=outcome, kind='bar')
[3]:
<Axes: xlabel='v0'>
[4]:
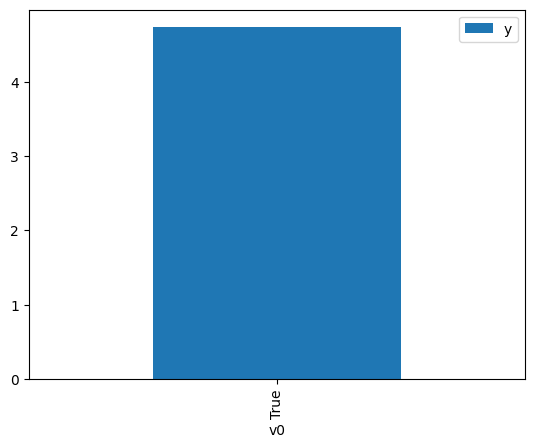
df.causal.do(x={treatment: 1},
variable_types={treatment:'b', outcome: 'c', common_cause: 'c'},
outcome=outcome,
method='weighting',
common_causes=[common_cause]
).groupby(treatment).mean().plot(y=outcome, kind='bar')
[4]:
<Axes: xlabel='v0'>
[5]:
cdf_1 = df.causal.do(x={treatment: 1},
variable_types={treatment: 'b', outcome: 'c', common_cause: 'c'},
outcome=outcome,
graph=nx_graph
)
cdf_0 = df.causal.do(x={treatment: 0},
variable_types={treatment: 'b', outcome: 'c', common_cause: 'c'},
outcome=outcome,
graph=nx_graph
)
[6]:
cdf_0
[6]:
| W0 | v0 | y | propensity_score | weight | |
|---|---|---|---|---|---|
| 0 | 2.404146 | False | 3.487173 | 0.221540 | 4.513856 |
| 1 | -0.119822 | False | 0.453772 | 0.542849 | 1.842132 |
| 2 | -0.614196 | False | 0.697349 | 0.611026 | 1.636591 |
| 3 | -0.064320 | False | 1.124111 | 0.535044 | 1.869006 |
| 4 | -0.137764 | False | -1.384972 | 0.545368 | 1.833623 |
| ... | ... | ... | ... | ... | ... |
| 995 | -0.776681 | False | -1.094488 | 0.632648 | 1.580659 |
| 996 | 0.397365 | False | 0.620665 | 0.469811 | 2.128515 |
| 997 | 0.713727 | False | 2.663563 | 0.425568 | 2.349803 |
| 998 | 1.595413 | False | 2.558598 | 0.310243 | 3.223276 |
| 999 | 1.712484 | False | 2.700458 | 0.296245 | 3.375582 |
1000 rows × 5 columns
[7]:
cdf_1
[7]:
| W0 | v0 | y | propensity_score | weight | |
|---|---|---|---|---|---|
| 0 | -0.607244 | True | 3.065915 | 0.389909 | 2.564699 |
| 1 | -0.777414 | True | 4.223191 | 0.367256 | 2.722898 |
| 2 | -2.025207 | True | 2.215251 | 0.222658 | 4.491188 |
| 3 | -1.701738 | True | 1.773618 | 0.255943 | 3.907120 |
| 4 | -0.578541 | True | 5.247765 | 0.393781 | 2.539485 |
| ... | ... | ... | ... | ... | ... |
| 995 | 0.044282 | True | 5.576950 | 0.480276 | 2.082137 |
| 996 | -2.149224 | True | 1.854231 | 0.210746 | 4.745046 |
| 997 | -0.777414 | True | 4.223191 | 0.367256 | 2.722898 |
| 998 | 1.066502 | True | 5.893722 | 0.622371 | 1.606758 |
| 999 | -0.543172 | True | 3.854451 | 0.398569 | 2.508973 |
1000 rows × 5 columns
Comparing the estimate to Linear Regression
First, estimating the effect using the causal data frame, and the 95% confidence interval.
[8]:
(cdf_1['y'] - cdf_0['y']).mean()
[8]:
$\displaystyle 5.03222846738008$
[9]:
1.96*(cdf_1['y'] - cdf_0['y']).std() / np.sqrt(len(df))
[9]:
$\displaystyle 0.125764811665961$
Comparing to the estimate from OLS.
[10]:
model = OLS(np.asarray(df[outcome]), np.asarray(df[[common_cause, treatment]], dtype=np.float64))
result = model.fit()
result.summary()
[10]:
| Dep. Variable: | y | R-squared (uncentered): | 0.929 |
|---|---|---|---|
| Model: | OLS | Adj. R-squared (uncentered): | 0.928 |
| Method: | Least Squares | F-statistic: | 6487. |
| Date: | Mon, 25 Dec 2023 | Prob (F-statistic): | 0.00 |
| Time: | 07:24:16 | Log-Likelihood: | -1372.1 |
| No. Observations: | 1000 | AIC: | 2748. |
| Df Residuals: | 998 | BIC: | 2758. |
| Df Model: | 2 | ||
| Covariance Type: | nonrobust |
| coef | std err | t | P>|t| | [0.025 | 0.975] | |
|---|---|---|---|---|---|---|
| x1 | 1.0182 | 0.028 | 36.171 | 0.000 | 0.963 | 1.073 |
| x2 | 5.0284 | 0.046 | 108.991 | 0.000 | 4.938 | 5.119 |
| Omnibus: | 2.186 | Durbin-Watson: | 1.932 |
|---|---|---|---|
| Prob(Omnibus): | 0.335 | Jarque-Bera (JB): | 2.056 |
| Skew: | 0.045 | Prob(JB): | 0.358 |
| Kurtosis: | 2.797 | Cond. No. | 1.64 |
Notes:
[1] R² is computed without centering (uncentered) since the model does not contain a constant.
[2] Standard Errors assume that the covariance matrix of the errors is correctly specified.